Analysis in relationships of overlapping genes in systemic sclerosis-associated interstitial lung disease, idiopathic pulmonary fibrosis and pulmonary fibrosis model induced by transforming growth factor-β
-
摘要:目的
利用生物信息学技术分析系统性硬化症相关间质性肺病(SSc-ILD)、特发性肺纤维化(IPF)与转化生长因子-β(TGF-β)诱导的肺纤维化模型重叠基因的相关性。
方法通过Gene Expression Omnibus(GEO)数据库获取SSc-ILD、IPF与TGF-β诱导的肺纤维化模型相关基因芯片数据GSE76808、GSE135099。采用R软件对数据进行分析,按照adj.P<0.05和|logFC|>1进行筛选,将筛选得到的基因进行基因交互分析;利用R软件对重叠基因进行基因本体(GO)功能注释、京都基因与基因组百科全书(KEGG)通路富集及可视化;基于String数据库对重叠基因进行蛋白质互作(PPI)分析;应用在线分析工具TISIDB获得hub基因并分析其功能。
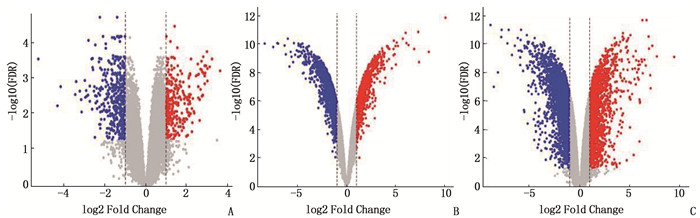
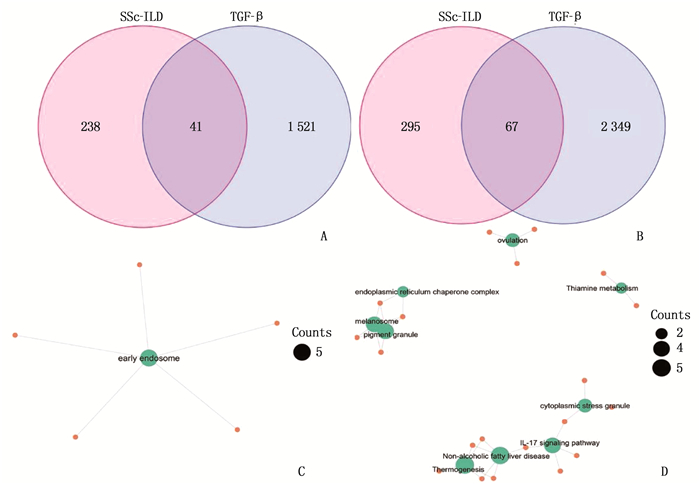
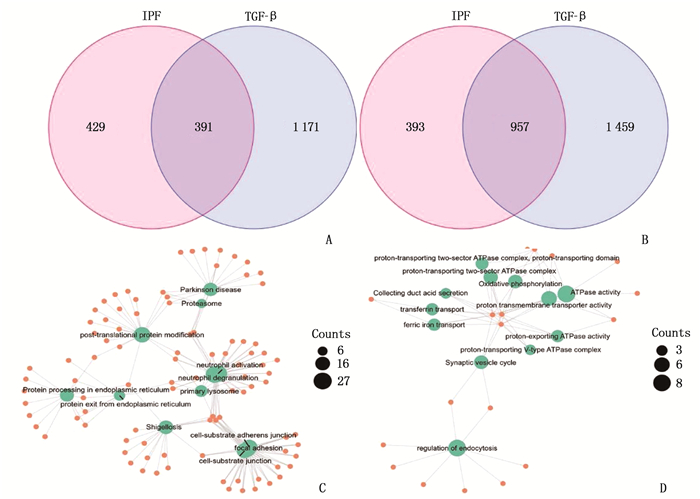
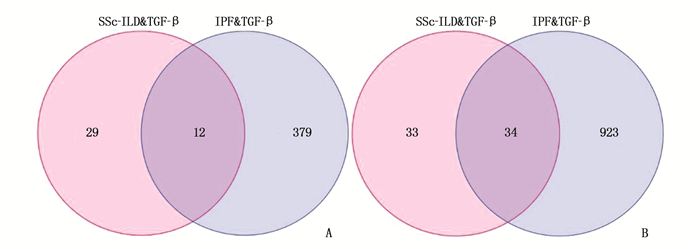
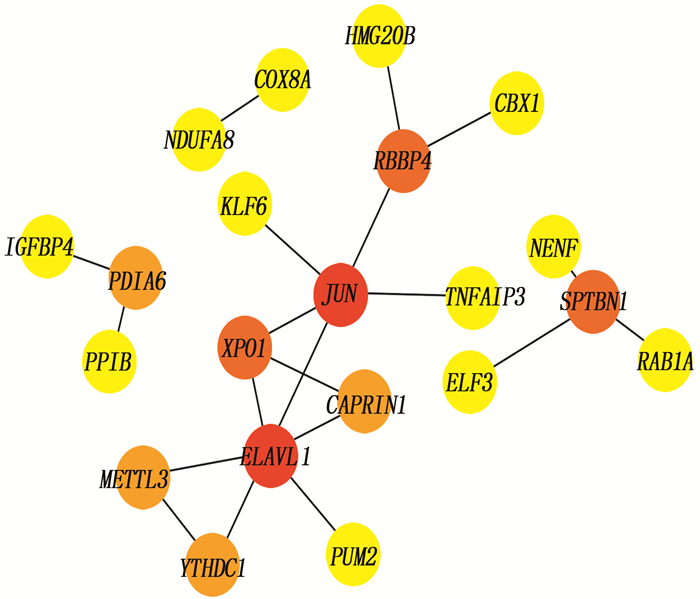
结果与健康对照组(NC组)相比,SSc-ILD、IPF、TGF-β诱导的肺纤维化模型中存在差异性表达的基因。在SSc-ILD、IPF、TGF-β诱导的肺纤维化模型均下调的34个重叠基因中存在PPI并获得hub基因ELAVL1。GO富集分析结果表明,ELAVL1基因不仅参与RNA剪接、加工、翻译等多种转录后修饰过程,还在腺苷酸活化蛋白激酶(AMPK)信号通路中存在富集。
结论在SSc-ILD和IPF两种疾病中,AMPK信号可能存在异常,导致TGF-β产生增多,而TGF-β可能通过调控ELAVL1蛋白(HuR)由胞核向胞浆转位,进而参与肺纤维化的调节,但具体机制仍有待进一步验证。
-
关键词:
- 转化生长因子-β /
- 系统性硬化症相关间质性肺病 /
- 特发性肺纤维化 /
- 生物信息学分析
Abstract:ObjectiveTo analyze the relationships of overlapping genes in systemic sclerosis-associated interstitial lung disease (SSc-ILD), idiopathic pulmonary fibrosis (IPF) and pulmonary fibrosis model induced by transforming growth factor-β (TGF-β) by using bioinformatics technology.
MethodsRelevant gene chip data GSE76808 and GSE135099 of SSc-ILD, IPF and pulmonary fibrosis model induced by TGF-β were obtained from Gene Expression Omnibus (GEO) database. R software was used to analyze the data, and according to the screening conditions of adj. P<0.05 and |logFC|>1, the screened genes were analyzed by gene interaction; Gene Ontology (GO) function annotation, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment and visualization for overlapping genes were performed by using R software; protein-protein interaction (PPI) analysis of overlapping genes was performed based on string database; the hub gene was obtained by analysis tool TISIDB and its function was analyzed.
ResultsCompared with the normal control group (NC group), there were differentially expressed genes in SSc-ILD, IPF and pulmonary fibrosis model induced by TGF-β. There was PPI existed in 34 overlapping genes down-regulated by SSc-ILD, IPF and pulmonary fibrosis model induced by TGF-β, and hub gene ELAVL1 was obtained. GO enrichment analysis results showed that ELAVL1 gene was not only involved in many post-transcriptional modification processes such as RNA splicing, processing and translation, but also enriched in adenylate activated protein kinase (AMPK) signal pathway.
ConclusionIn SSc-ILD and IPF, the AMPK signal may be abnormal, resulting in increasing of TGF-β, and TGF-β may be involved in the regulation of pulmonary fibrosis by regulating the translocation of ELAVL1 protein (HuR) from nucleus to cytoplasm, but the specific mechanism still needs to be further verified.
-
良性前列腺增生(BPH)是因前列腺组织良性增大引发的下尿路梗阻的泌尿系统疾病,多发于中老年男性,严重影响患者身心健康[1-2]。目前, BPH发病相关因素尚未完全阐明,研究[3-4]认为BPH发病与高水平雄激素、年龄、高血糖、高血压、高脂血症及炎症相关。既往研究[5-7]显示,前列腺特异性抗原(PSA)水平异常与前列腺癌、前列腺炎、前列腺良性增生等发生发展密切相关。游离前列腺特异性抗原(fPSA)是总前列腺特异性抗原(tPSA)游离形式,约占血液tPSA的30%, 其在BPH患者血清中呈高水平表现,可能参与并影响BPH发病过程[8]。经尿道前列腺电切术是BPH患者手术治疗的金标准,但术后可引起尿失禁、电切综合征等并发症[9-10], 且手术切除不彻底,临床应用受到限制。斯钦布和等[11]发现经尿道前列腺剜除术(TUERP)的术后并发症较少,可有效清除良性前列腺增生组织,恢复尿道功能,改善前列腺增生症状。TUERP虽可改善BPH患者生存质量,但治疗后仍存在复发和预后不良的情况[12]。本研究检测BPH患者TUERP术后血清fPSA的水平,分析fPSA与前列腺体积的关系,探讨BPH患者术后预后情况,现将结果报告如下。
1. 资料与方法
1.1 临床资料
选取2017年1月—2020年6月行TUERP的BPH患者105例为研究对象。参考直肠超声所得前列腺体积将BPH患者分为小体积组(n=49)、中体积组(n=36)、大体积组(n=20)。小体积组年龄54~70岁,平均(61.45±6.78)岁; 前列腺体积 < 40 mL, 平均体积(31.22±8.63) mL。中体积组年龄53~71岁,平均(61.89±6.90)岁; 前列腺体积40~80 mL, 平均体积(60.13±9.25) mL。大体积组年龄54~72岁,平均(62.52±6.97)岁; 前列腺体积>80 mL, 平均体积(90.56±9.78) mL。3组年龄比较,差异无统计学意义(P>0.05)。收集BPH患者甘油三酯(TG)、高密度脂蛋白胆固醇(HDL-C)、血糖、高血压病、吸烟史、饮酒史等临床资料。
诊断标准: 参考《良性前列腺增生诊断治疗指南》[13]有关BPH判定标准进行诊断。纳入标准: ①患者均符合BPH相关诊断标准,结合尿常规检查、影像学检查、病史、内镜检查、尿动力学检查确诊; ②患者均自愿参与本研究。排除标准: ①合并凝血功能异常者; ②复发BPH者; ③手术治疗不能耐受者; ④随访资料不全者。本研究符合《赫尔辛基宣言》相关要求,获得伦理委员会批准。
1.2 方法
采用人fPSA化学发光检测试剂盒(货号314381)以及全自动化学发光免疫分析仪(型号LIAISON-XL), 均购自意大利索灵公司。采集所有患者术前及术后1、3、7 d的静脉血4~5 mL, 静置25~35 min, 4 450转/min离心6 min, 取上清液,保存于-80 ℃冰箱中待检。取出冻存血清样本并解冻,严格按照fPSA ELISA试剂盒说明书检测3组受试者血清fPSA水平[14]。
105例BPH患者均接受TUERP, 进行6个月随访,应用国际前列腺症状评分(IPSS)评估患者症状改善情况[7]。IPSS评分为0~35分,其中0~7分、8~19分、20~35分依次表示轻、中、重度症状,将IPSS评分≥8分及BPH复发患者设为预后不良组, IPSS评分 < 8分患者设为预后良好组。
1.3 统计学方法
本研究所有数据采用SPSS 25.0软件进行统计分析。符合正态分布的计量数据以均数±标准差(x±s)表示,组间比较采用独立样本t检验; 计数资料以(n)表示,行卡方检验。Pearson法分析BPH患者术前血清fPSA水平与前列腺体积的相关性。Logistic回归分析法分析BPH患者预后的影响因素。P < 0.05为差异有统计学意义。
2. 结果
2.1 不同前列腺体积组患者血清fPSA水平比较
术前及术后1 d时,小体积组、中体积组、大体积组患者血清fPSA水平呈增高趋势,组间比较差异均有统计学意义(P < 0.05), 见表 1。
表 1 不同前列腺体积组患者血清fPSA水平比较(x±s)ng/mL 组别 n 术前 术后1 d 术后3 d 术后7 d 小体积组 49 1.37±0.56 2.49±1.05 2.14±1.01 2.05±0.87 中体积组 36 4.68±1.93* 6.54±2.37* 2.60±1.16 1.97±0.83 大体积组 20 7.06±2.67*# 8.42±3.65*# 2.61±1.18 1.99±0.84 fPSA: 游离前列腺特异性抗原。与小体积组比较, *P < 0.05; 与中体积组比较, #P < 0.05。 2.2 BPH患者术前血清fPSA水平与前列腺体积的关系
Pearson法分析结果显示, BPH患者术前血清fPSA水平与前列腺体积呈正相关(r=0.598, P < 0.05)。见图 1。
2.3 BPH患者复发和预后不良的情况
105例BPH患者均随访6个月,复发患者5例(4.76%)。中体积组复发4例,其中1例于术后3个月复发, 3例于术后4个月复发; 3例患者当月再次接受手术治疗,预后良好, 1例患者在随访期间未接受再次治疗。大体积组复发1例,该例患者于术后4个月复发,当月再次进行手术治疗,预后良好。5例复发患者均不统计其在随访6个月时IPSS得分,但将其纳入不良预后组。其余100例BPH患者术后6个月IPSS得分为(4.72±1.68)分,其中21例IPSS得分≥8分,故预后不良组患者共26例(24.76%)。与预后良好组(n=79)相比,预后不良组患者HDL-C、血糖、术后7 d血清fPSA水平及合并高血压比率均升高,差异有统计学意义(P < 0.05), 见表 2。
表 2 不同预后BPH患者临床资料及术后7 d血清fPSA水平比较(x±s)指标 预后良好组(n=79) 预后不良组(n=26) χ2/t P 年龄/岁 61.86±9.63 62.28±9.92 0.191 0.849 TG ≥1.7 mmol/L 28 9 0.006 0.939 < 1.7 mmol/L 51 17 HDL-C ≥1.03 mmol/L 8 7 4.507 0.034 < 1.03 mmol/L 71 19 血糖 ≥6.2 mmol/L 20 15 7.635 0.006 < 6.2 mmol/L 59 11 合并高血压 是 19 14 8.058 0.005 否 60 12 吸烟史 是 40 13 0.003 0.955 否 39 13 饮酒史 是 42 14 0.004 0.952 否 37 12 术后7 d血清fPSA水平/(ng/mL) 1.66±0.77 3.38±1.45 7.767 < 0.001 TG: 甘油三酯; HDL-C: 高密度脂蛋白胆固醇; fPSA: 游离前列腺特异性抗原。 2.4 Logistic回归分析BPH患者预后的影响因素
以BPH患者预后为因变量,以BPH患者HDL-C、血糖水平、合并高血压、术后7 d血清fPSA水平为自变量,进行Logistic回归分析,结果显示HDL-C、血糖、合并高血压、术后7 d血清fPSA均为BPH患者预后的危险因素(P < 0.05)。见表 3。
表 3 BPH患者预后的影响因素的Logistic回归分析指标 B SE Wald P OR 95%CI HDL-C 0.579 0.218 7.051 0.008 1.784 1.164~2.735 血糖 0.516 0.212 5.934 0.015 1.676 1.106~2.539 合并高血压 0.692 0.231 8.965 0.003 1.997 1.270~3.141 术后7 d血清fPSA 0.616 0.224 7.569 0.006 1.852 1.194~2.873 HDL-C: 高密度脂蛋白胆固醇; fPSA: 游离前列腺特异性抗原。 3. 讨论
随着全球人口老龄化的加剧, BPH发病率呈上升趋势,其以尿急、尿频、夜尿频繁及排尿困难为主要临床表现,可引起血尿、膀胱结石、泌尿系统感染等并发症,部分患者可发生恶变,严重影响患者生活质量[15-17]。有学者[18-19]根据前列腺开放摘除术原理,在前列腺分隔式切除基础上创建了TUERP, 该术式在内镜下从外科包膜剥离前列腺,利用组织粉碎器取出增生组织,从而达到治疗BPH的目的。TUERP是临床治疗BPH的常用术式,但TUERP后因初次手术时增生腺体切除不彻底,易导致术后残余的腺体再次增生,并引发膀胱出口梗阻,导致BPH复发。
PSA是由前列腺上皮细胞分泌产生的一种特异性糖蛋白,存在于精液和前列腺组织中,多数PSA产生后随精液排出体外,且前列腺泡、导管腔和血液系统间存在组织屏障,在正常情况下,血清PSA含量较低[20-21]。当处于病理状态下时,淋巴系统与前列腺泡组织间的屏障被破坏,外周血PSA水平异常升高,有助于筛查前列腺疾病[22]。研究[23]显示, PSA在前列腺癌中表达水平明显升高,其可辅助诊断和评估前列腺癌。尿液PSA水平可诊断并预测BPH的严重程度,且其水平随炎症分级升高及侵袭程度加重而呈上升趋势[24-25], 表明异常水平的PSA与BPH类前列腺疾病关系密切。蒋团建等[26]对92例经尿道前列腺等离子双极电切术的BPH患者进行研究,结果显示,术后7 d血清PSA水平预测BPH患者不良预后的曲线下面积为0.823, 灵敏度、特异度分别为83.3%、66.2%。
COBAN S等[27]对1 000例BPH患者进行回顾性研究,结果显示, BPH患者的年龄、tPSA、fPSA与前列腺体积显著相关,其中fPSA预测前列腺体积的价值更高。本研究结果显示,术后1 d时,不同前列腺体积患者的fPSA均处于较高水平,提示fPSA可能与BPH疾病进展密切相关,原因可能是TUERP破坏了前列腺,使PSA释放入血,引起血清fPSA水平升高。本研究还显示,术前及术后1 d时,小体积组、中体积组、大体积组患者血清fPSA水平呈增高趋势,组间比较差异均有统计学意义(P < 0.05), 原因可能是TUERP彻底清除前列腺增生中央带和移行带,仅前列腺外周带被保留,故所有BPH患者的fPSA生成源体积相似,因此fPSA生成量及释放入血量大致相同。此外,小体积组BPH患者接受TUERP前,其前列腺组织损伤相对较轻,引发PSA释放入血量相应较少,术后7 d时因手术引起前列腺损伤,导致PSA释放入血量较高,故小体积组BPH患者在术后7 d时血清fPSA水平较术前升高。
本研究结果显示, BPH患者术前血清fPSA水平与前列腺体积呈正相关,与COBAN S等[27]研究结果相似,提示fPSA可能与前列腺体积相互影响,进而影响BPH发生发展。本研究中BPH患者术后血清fPSA水平逐渐下降并趋于稳定,提示血清fPSA水平在术后早期存在一个较稳定的低值,如果血清fPSA超过该值,提示增生腺体切除不彻底或患者合并其他引起fPSA升高的疾病。本研究还显示,术后7 d的血清fPSA水平低于术后3 d时,表明术后3~7 d的血清fPSA水平呈下降趋势,故选择术后7 d血清fPSA水平评估BPH患者预后情况。本研究只选取了术后3个时间点,无法确定术后7 d是最佳检测及预测BPH患者预后的时间点,有待后续研究证实。本研究中BPH患者复发率、预后不良率分别为4.76%、24.76%, 且预后不良组患者HDL-C、血糖、术后7 d血清fPSA水平及合并高血压比率均显著高于预后良好组,提示BPH患者TUERP后存在复发和预后不良的情况。进一步研究发现, HDL-C、血糖、合并高血压、术后7 d血清fPSA均是BPH患者预后的危险因素,提示HDL-C、血糖、术后7 d血清fPSA水平升高均会增高BPH患者发生不良预后的风险。
综上所述, fPSA在BPH患者血清中呈高水平表现,其可能参与影响BPH发病过程,术后7 d血清fPSA水平可评估BPH患者TUERP后的预后情况。
-
表 1 PPI网络分析中前21个基因
排序 名称 得分 排序 名称 得分 1 ELAVL1 6 10 RAB1A 1 2 JUN 5 10 COX8A 1 3 RBBP4 3 10 NDUFA8 1 3 SPTBN1 3 10 PUM2 1 3 XPO1 3 10 ELF3 1 6 METTL3 2 10 HMG20B 1 6 YTHDC1 2 10 IGFBP4 1 6 PDIA6 2 10 TNFAIP3 1 6 CAPRIN1 2 10 KLF6 1 10 PPIB 1 10 NENF 1 10 CBX1 1 表 2 GO分析 ELAVL1 基因
GO号 生物过程 GO号 生物过程 GO号 生物过程 0000375 通过酯交换反应进行RNA剪接 0031047 RNA基因沉默 0048255 mRNA稳定 0000377 通过酯交换反应与凸起的腺苷作为亲核试剂进行RNA剪接 0034248 细胞内酰胺代谢过程的调节 0060147 转录后基因沉默的调控 0000398 通过剪接体进行mRNA剪接 0034249 细胞内酰胺代谢过程的负调控 0060149 转录后基因沉默的负调控 0006397 mRNA加工 0034250 细胞内酰胺代谢过程的正向调控 0060964 miRNA对基因沉默的调控 0006417 调节翻译 0035194 RNA转录后基因沉默 0060965 miRNA对基因沉默的负调控 0008380 RNA剪接 0035195 miRNA的基因沉默 0060966 RNA对基因沉默的调控 0010608 基因表达的转录后调控 0040029 基因表达调控、表观遗传 0060967 RNA对基因沉默的负调控 0016441 转录后基因沉默 0043487 RNA稳定性的调节 0060968 基因沉默的调控 0016458 基因沉默 0043488 mRNA稳定性的调节 0060969 基因沉默的负调控 0017148 翻译的负面调节 0043489 RNA稳定 0070935 3′-UTR介导的mRNA稳定性 0019827 维持干细胞群 0045727 正向调节翻译 0098727 维持细胞数量 表 3 GO分析 ELAVL1 基因
GO号 分子功能 0003725 结合双链RNA 0003729 结合mRNA 0003730 结合mRNA 3′-UTR 0017091 结合富含AU元素 0035925 结合mRNA 3′-UTR富含AU元素区域 -
[1] ZHANG X L, YUN J S, HAN D, et al. TGF-β pathway in salivary gland fibrosis[J]. Int J Mol Sci, 2020, 21(23): E9138. doi: 10.3390/ijms21239138
[2] MORIKAWA M, DERYNCK R, MIYAZONO K. TGF-β and the TGF-β family: context-dependent roles in cell and tissue physiology[J]. Cold Spring Harb Perspect Biol, 2016, 8(5): a021873. doi: 10.1101/cshperspect.a021873
[3] HATA A, CHEN Y G. TGF-β signaling from receptors to smads[J]. Cold Spring Harb Perspect Biol, 2016, 8(9): a022061. doi: 10.1101/cshperspect.a022061
[4] LAMOUILLE S, XU J, DERYNCK R. Molecular mechanisms of epithelial-mesenchymal transition[J]. Nat Rev Mol Cell Biol, 2014, 15(3): 178-196. doi: 10.1038/nrm3758
[5] TRIPATHI V, SIXT K M, GAO S J, et al. Direct regulation of alternative splicing by SMAD3 through PCBP1 is essential to the tumor-promoting role of TGF-Β[J]. Mol Cell, 2016, 64(3): 549-564. doi: 10.1016/j.molcel.2016.09.013
[6] BUDI E H, DUAN D N, DERYNCK R. Transforming growth factor-β receptors and smads: regulatory complexity and functional versatility[J]. Trends Cell Biol, 2017, 27(9): 658-672. doi: 10.1016/j.tcb.2017.04.005
[7] MENG X M, NIKOLIC-PATERSON D J, LAN H Y. TGF-β: the master regulator of fibrosis[J]. Nat Rev Nephrol, 2016, 12(6): 325-338. doi: 10.1038/nrneph.2016.48
[8] ELHAI M, MEUNE C, BOUBAYA M, et al. Mapping and predicting mortality from systemic sclerosis[J]. Ann Rheum Dis, 2017, 76(11): 1897-1905. doi: 10.1136/annrheumdis-2017-211448
[9] KHEDOE P, MARGES E, HIEMSTRA P, et al. Interstitial lung disease in patients with systemic sclerosis: toward personalized-medicine-based prediction and drug screening models of systemic sclerosis-related interstitial lung disease (SSc-ILD)[J]. Front Immunol, 2020, 11: 1990. doi: 10.3389/fimmu.2020.01990
[10] PERELAS A, SILVER R M, ARROSSI A V, et al. Systemic sclerosis-associated interstitial lung disease[J]. Lancet Respir Med, 2020, 8(3): 304-320. doi: 10.1016/S2213-2600(19)30480-1
[11] DENTON C P, KHANNA D. Systemic sclerosis[J]. Lancet, 2017, 390(10103): 1685-1699. doi: 10.1016/S0140-6736(17)30933-9
[12] CABRAL-MARQUES O, RIEMEKASTEN G. Vascular hypothesis revisited: role of stimulating antibodies against angiotensin and endothelin receptors in the pathogenesis of systemic sclerosis[J]. Autoimmun Rev, 2016, 15(7): 690-694. doi: 10.1016/j.autrev.2016.03.005
[13] LAURENT P, ALLARD B, MANICKI P, et al. TGFβ promotes low IL10-producing ILC2 with profibrotic ability involved in skin fibrosis in systemic sclerosis[J]. Ann Rheum Dis, 2021, 80(12): 1594-1603. doi: 10.1136/annrheumdis-2020-219748
[14] HSU W L, HSIEH Y C, YU H S, et al. 2-Aminoethyl diphenylborinate inhibits bleomycin-induced skin and pulmonary fibrosis via interrupting intracellular Ca2+ regulation[J]. J Dermatol Sci, 2021, 103(2): 101-108. doi: 10.1016/j.jdermsci.2021.07.005
[15] GEORGE P M, PATTERSON C M, REED A K, et al. Lung transplantation for idiopathic pulmonary fibrosis[J]. Lancet Respir Med, 2019, 7(3): 271-282. doi: 10.1016/S2213-2600(18)30502-2
[16] RICHELDI L, COLLARD H R, JONES M G. Idiopathic pulmonary fibrosis[J]. Lancet, 2017, 389(10082): 1941-1952. doi: 10.1016/S0140-6736(17)30866-8
[17] AL-TAMARI H M, DABRAL S, SCHMALL A, et al. FoxO3 an important player in fibrogenesis and therapeutic target for idiopathic pulmonary fibrosis[J]. EMBO Mol Med, 2018, 10(2): 276-293. doi: 10.15252/emmm.201606261
[18] HOSSEINZADEH A, JAVAD-MOOSAVI S A, REITER R J, et al. Idiopathic pulmonary fibrosis (IPF) signaling pathways and protective roles of melatonin[J]. Life Sci, 2018, 201: 17-29. doi: 10.1016/j.lfs.2018.03.032
[19] KOLB M, BONELLA F, WOLLIN L. Therapeutic targets in idiopathic pulmonary fibrosis[J]. Respir Med, 2017, 131: 49-57. doi: 10.1016/j.rmed.2017.07.062
[20] EDGAR R, DOMRACHEV M, LASH A E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository[J]. Nucleic Acids Res, 2002, 30(1): 207-210. doi: 10.1093/nar/30.1.207
[21] SZKLARCZYK D, MORRIS J H, COOK H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible[J]. Nucleic Acids Res, 2017, 45(D1): D362-D368. doi: 10.1093/nar/gkw937
[22] RU B B, WONG C N, TONG Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions[J]. Bioinformatics, 2019, 35(20): 4200-4202. doi: 10.1093/bioinformatics/btz210
[23] STOCK C J W, MICHAELOUDES C, LEONI P, et al. Bromodomain and extraterminal (BET) protein inhibition restores redox balance and inhibits myofibroblast activation[J]. Biomed Res Int, 2019, 2019: 1484736.
[24] VALENZI E, TABIB T, PAPAZOGLOU A, et al. Disparate interferon signaling and shared aberrant basaloid cells in single-cell profiling of idiopathic pulmonary fibrosis and systemic sclerosis-associated interstitial lung disease[J]. Front Immunol, 2021, 12: 595811. doi: 10.3389/fimmu.2021.595811
[25] DECARIS M L, SCHAUB J R, CHEN C, et al. Dual inhibition of αvβ6 and αvβ1 reduces fibrogenesis in lung tissue explants from patients with IPF[J]. Respir Res, 2021, 22(1): 265. doi: 10.1186/s12931-021-01863-0
[26] DISTLER J H W, GYÖRFI A H, RAMANUJAM M, et al. Shared and distinct mechanisms of fibrosis[J]. Nat Rev Rheumatol, 2019, 15(12): 705-730. doi: 10.1038/s41584-019-0322-7
[27] ANGIOLILLI C, MARUT W, VAN DER KROEF M, et al. New insights into the genetics and epigenetics of systemic sclerosis[J]. Nat Rev Rheumatol, 2018, 14(11): 657-673. doi: 10.1038/s41584-018-0099-0
[28] TRUCHETET M E, BREMBILLA N C, CHIZZOLINI C. Current concepts on the pathogenesis of systemic sclerosis[J]. Clin Rev Allergy Immunol, 2021: 1-13.
[29] ALLEN R J, GUILLEN-GUIO B, OLDHAM J M, et al. Genome-wide association study of susceptibility to idiopathic pulmonary fibrosis[J]. Am J Respir Crit Care Med, 2020, 201(5): 564-574. doi: 10.1164/rccm.201905-1017OC
[30] LORENZO-SALAZAR J M, MA S F, JOU J, et al. Novel idiopathic pulmonary fibrosis susceptibility variants revealed by deep sequencing[J]. ERJ Open Res, 2019, 5(2): 00071-02019.
[31] WANG J, GUO Y, CHU H L, et al. Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis[J]. Int J Mol Sci, 2013, 14(5): 10015-10041. doi: 10.3390/ijms140510015
[32] ZHANG F, CAI Z L, LV H D, et al. Multiple functions of HuR in urinary tumors[J]. J Cancer Res Clin Oncol, 2019, 145(1): 11-18. doi: 10.1007/s00432-018-2778-2
[33] WOODHOO A, IRUARRIZAGA-LEJARRETA M, BERAZA N, et al. Human antigen R contributes to hepatic stellate cell activation and liver fibrosis[J]. Hepatology, 2012, 56(5): 1870-1882. doi: 10.1002/hep.25828
[34] AL-HABEEB F, ALOUFI N, TRABOULSI H, et al. Human antigen R promotes lung fibroblast differentiation to myofibroblasts and increases extracellular matrix production[J]. J Cell Physiol, 2021, 236(10): 6836-6851. doi: 10.1002/jcp.30380
[35] GAO L, WANG L Y, LIU Z Q, et al. TNAP inhibition attenuates cardiac fibrosis induced by myocardial infarction through deactivating TGF-β1/Smads and activating P53 signaling pathways[J]. Cell Death Dis, 2020, 11(1): 44. doi: 10.1038/s41419-020-2243-4
[36] XIAO Y C, YE J T, ZHOU Y, et al. Baicalin inhibits pressure overload-induced cardiac fibrosis through regulating AMPK/TGF-β/Smads signaling pathway[J]. Arch Biochem Biophys, 2018, 640: 37-46. doi: 10.1016/j.abb.2018.01.006
[37] ZHANG P J, CAO M F, LIU Y, et al. PDGF-induced airway smooth muscle proliferation is associated with Human antigen R activation and could be weakened by AMPK activation[J]. Mol Biol Rep, 2012, 39(5): 5819-5829. doi: 10.1007/s11033-011-1392-z
-
期刊类型引用(2)
1. 喻洁明,任荣飞,陈晓亮. 耳内镜下应用耳屏软骨-软骨膜复合体行Ⅰ型鼓室成形术的临床疗效研究. 中国医学文摘(耳鼻咽喉科学). 2024(01): 130-132+108 .  百度学术
百度学术
2. 包勇正,邹文兰,胡志邦,马敬. 29例鼓膜大穿孔患者耳内镜下鼓膜成形术疗效观察. 实用临床医药杂志. 2020(23): 42-44 .  本站查看
本站查看
其他类型引用(0)




 下载:
下载:

 苏公网安备 32100302010246号
苏公网安备 32100302010246号
