Analysis in expression profiles of circular RNA in plasma in children with pediatric type 2 spinal muscular atrophy
-
摘要:目的
分析儿童脊髓性肌萎缩症(SMA)2型患者血浆中环状RNA (circRNA)的表达谱变化。
方法选取2021年9月—2022年3月安徽省儿童医院神经内科住院治疗的儿童SMA2型患者5例为SMA组, 同期健康对照者5例为对照组。采用高通量测序技术检测并筛选出血浆中差异表达的circRNA, 应用生物信息学进行基因本体论(GO)注释、京都基因和基因组百科全书(KEGG)和Reactome通路富集分析。利用在线数据库预测circRNA可能靶向的微小RNA (miRNA)。
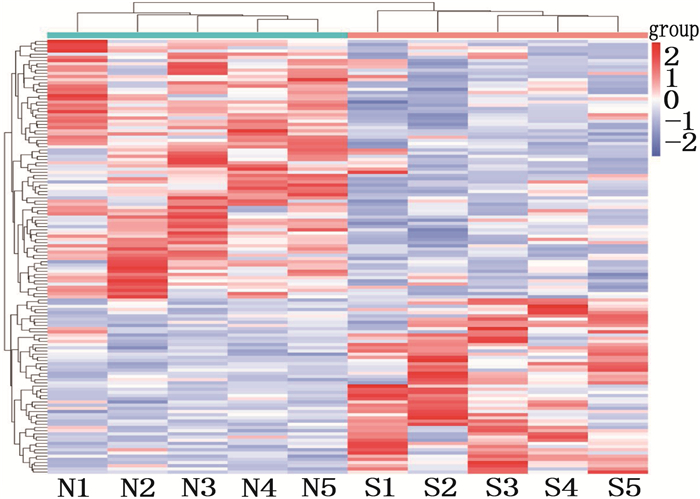
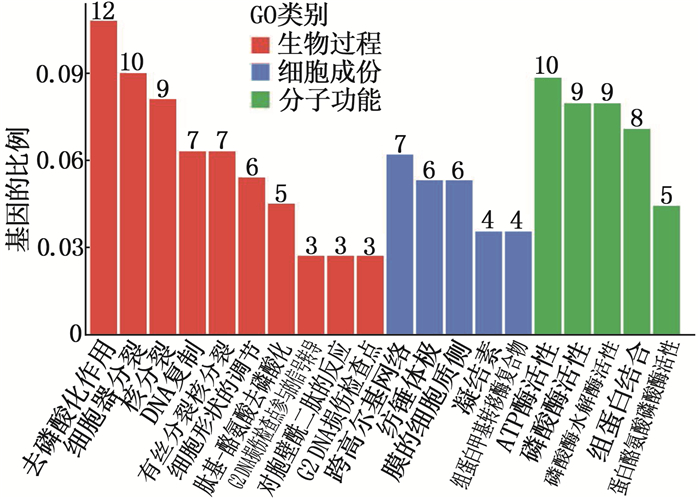
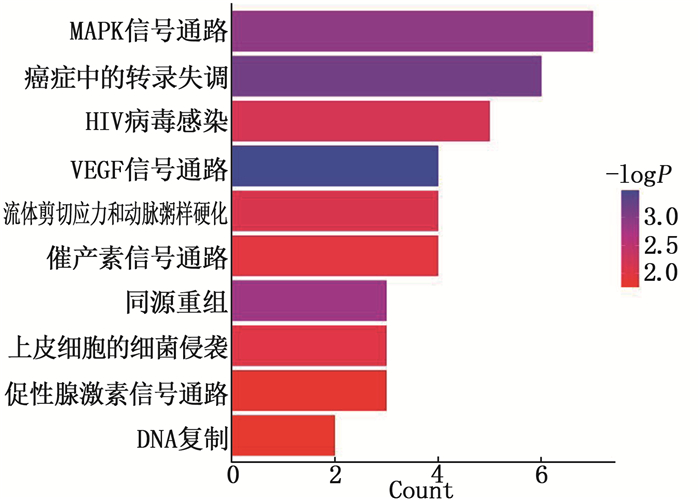
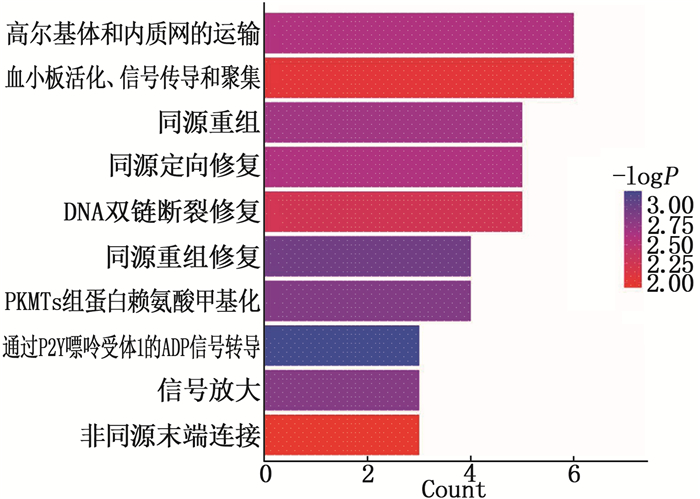
结果与对照组相比, SMA组患者血浆中共有136个circRNA呈显著差异性表达(P < 0.05, 差异倍数≥1.5), 包括55个表达上调和81个表达下调的circRNA。生物信息学分析发现,同源重组修复、DNA复制等通路在SMA的发生发展中具有重要作用。应用TargetScan和miRanda软件预测了差异表达circRNA与miRNA的关系,绘制了circRNA-miRNA调控网络图。
结论SMA组与对照组存在差异表达的circRNA。这些circRNA可能参与SMA的发生、发展,或可成为SMA的新型诊断和治疗的潜在分子标志物。
Abstract:ObjectiveTo analyze the change of expression profiles of circular RNA (circRNA) in plasma of children with pediatric type 2 spinal muscular atrophy (SMA).
MethodsFrom September 2021 to March 2022, five hospitalized children with pediatric type 2 SMA in the Department of Neurology of Anhui Provincial Children's Hospital were selected as SMA group, and five healthy controls in the same period were selected as control group. High throughput sequencing technology was used to detect and screen the differentially expressed circRNA in plasma, and bioinformatics was used for gene ontology (GO) annotation, the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome pathway enrichment analyses. Online database was used to predict the microRNAs (miRNA) that circRNA may target.
ResultsCompared with the control group, a total of 136 circRNA were significantly differentially expressed in the plasma of children with SMA (P < 0.05, fold change≥1.5), including 55 up-regulated and 81 down-regulated circRNA. Bioinformatics analysis revealed that pathways such as homologous recombination repair and DNA replication play important roles in the occurrence and development of SMA. TargetScan and miRanda software were used to predict the relationship between differentially expressed circRNA and miRNA, and the picture of the circRNA-miRNA regulatory networks was drawn.
ConclusionThere are differentially expressed circRNA between the SMA group and the control group. These circRNA may be involved in the occurrence and development of SMA, and may become potential molecular markers for novel diagnosis and treatment of SMA in the future.
-
-
表 1 SMA组5例SMA患儿临床资料
临床资料 病例1 病例2 病例3 病例4 病例5 性别 女 男 女 男 女 就诊年龄 4岁9个月 3岁 10岁 3岁 12岁10个月 发病年龄 6个月 6个月 12个月 10个月 9个月 肌肉萎缩 + + + + + 骨骼畸形 + + + + + 肌张力 减弱 减弱 减弱 减弱 减弱 左肩-肘-腕-掌指肌力 4 3 2 2 3 右肩-肘-腕-掌指肌力 4 3 2 2 3 左髋-膝-踝-掌趾肌力 2 2 1 1 2 右髋-膝-踝-掌趾肌力 2 2 1 1 2 骨密度(胫骨中段) 0.6 1.3 -5.9 0.5 -1.8 表 2 SMA组与正常对照组差异表达的circRNA
环状RNA 染色体定位 基因名 表达情况 log2倍数变化 差异倍数 P hsa_circ_0024193 chr11 ATM 高 2.353 5.110 < 0.001 hsa_circ_0008833 chr6 SAMD3 高 2.098 4.281 0.001 hsa_circ_0000944 chr19 CCDC9 高 2.044 4.125 < 0.001 hsa_circ_0004137 chr14 RBM23 高 2.014 4.040 0.002 hsa_circ_0004751 chr17 AATF 高 1.934 3.821 0.007 hsa_circ_0005573 chr20 STK4 高 1.828 3.552 0.007 hsa_circ_0000615 chr15 ZNF609 高 1.812 3.511 < 0.001 hsa_circ_0007694 chr11 ATM 高 1.738 3.336 < 0.001 hsa_circ_0135062 chr7 ANKIB1 高 1.722 3.299 0.003 hsa_circ_0006956 chr10 CCSER2 高 1.709 3.269 0.004 hsa_circ_0005910 chr2 LTBP1 低 -2.624 0.162 < 0.001 hsa_circ_0009109 chr1 DCAF6 低 -2.320 0.200 0.002 hsa_circ_0076179 chr6 MAPK14 低 -2.084 0.235 0.003 hsa_circ_0028899 chr12 RNF10 低 -2.032 0.245 0.004 hsa_circ_0006809 chr14 CDC42BPB 低 -1.924 0.264 0.004 hsa_circ_0035957 chr15 DENND4A 低 -1.819 0.283 0.004 hsa_circ_0007017 chr7 KMT2C 低 -1.759 0.295 0.015 hsa_circ_0006272 chr10 CCAR1 低 -1.737 0.300 0.007 hsa_circ_0018403 chr10 SGMS1 低 -1.680 0.312 0.004 hsa_circ_0101926 chr14 KLHDC1 低 -1.665 0.315 0.015 表 3 差异表达的circRNA与miRNA结合位点的预测结果
表达趋势 环状RNA 结合相关miRNA hsa_circ_0024193 hsa-miR-513a-3p hsa-miR-548c-3p hsa-miR-507 hsa-miR-557 hsa-miR-324-5p 上调 hsa_circ_0008833 hsa-miR-1234 hsa-miR-586 hsa-miR-155 hsa-miR-616 hsa-miR-598 hsa_circ_0000944 hsa-miR-1307 hsa-miR-548b-3p hsa-miR-942 hsa-miR-136 hsa-miR-361-3p hsa_circ_0004137 hsa-miR-876-3p hsa-miR-31 hsa-miR-619 hsa-miR-1208 hsa-miR-1178 hsa_circ_0004751 hsa-miR-1208 hsa-miR-623 hsa-miR-766 hsa-miR-513a-3p hsa-miR-224 下调 hsa_circ_0005910 hsa-miR-1258 hsa-miR-184 hsa-miR-485-3p hsa-miR-488 hsa-miR-623 hsa_circ_0009109 hsa-miR-1256 hsa-miR-624 hsa-miR-577 hsa-miR-579 hsa-miR-557 hsa_circ_0076179 hsa-miR-769-5p hsa-miR-203 hsa-miR-516b hsa-miR-657 hsa-miR-1288 hsa_circ_0028899 hsa-miR-1248 hsa-miR-647 hsa-miR-194 hsa-miR-31 hsa-miR-183 hsa_circ_0006809 hsa-miR-766 hsa-miR-1205 hsa-miR-1248 hsa-miR-578 hsa-miR-576-5p -
[1] LI C, GENG Y, ZHU X, et al. The prevalence of spinal muscular atrophy carrier in China: evidences from epidemiological surveys[J]. Medicine (Baltimore), 2020, 99(5): e18975. doi: 10.1097/MD.0000000000018975
[2] EICHELBERGER E J, ALVES C R R, ZHANG R, et al. Increased systemic HSP70B levels in spinal muscular atrophy infants[J]. Ann Clin Transl Neurol, 2021, 8(7): 1495-1501. doi: 10.1002/acn3.51377
[3] MAASS P G, GLAŽAR P, MEMCZAK S, et al. A map of human circular RNAs in clinically relevant tissues[J]. J Mol Med (Berl), 2017, 95(11): 1179-1189. doi: 10.1007/s00109-017-1582-9
[4] KRISTENSEN L S, ANDERSEN M S, STAGSTED L V W, et al. The biogenesis, biology and characterization of circular RNAs[J]. Nat Rev Genet, 2019, 20(11): 675-691. doi: 10.1038/s41576-019-0158-7
[5] JIANG L, WANG X, ZHAN X, et al. Advance in circular RNA modulation effects of heart failure[J]. Gene, 2020, 763S: 100036.
[6] CHEN B, HUANG S. Circular RNA: An emerging non-coding RNA as a regulator and biomarker in cancer[J]. Cancer Letters, 2018, 418: 41-50. doi: 10.1016/j.canlet.2018.01.011
[7] GRAY L G, MILLS J D, CURRY-HYDE A, et al. Identification of Specific Circular RNA Expression Patterns and MicroRNA Interaction Networks in Mesial Temporal Lobe Epilepsy[J]. Front Genet, 2020, 11: 564301. doi: 10.3389/fgene.2020.564301
[8] CHEN D, HAO S, XU J. Revisiting the Relationship Between Alzheimer's Disease and Cancer With a circRNA Perspective[J]. Front Cell Dev Biol, 2021, 9: 647197. doi: 10.3389/fcell.2021.647197
[9] KUMAR L, SHAMSUZZAM A, JADIYA P, et al. Functional Characterization of Novel Circular RNA Molecule, circzip-2 and Its Synthesizing Gene zip-2 in C. elegans Model of Parkinson's Disease[J]. Mol Neurobiol, 2018, 55(8): 6914-6926. doi: 10.1007/s12035-018-0903-5
[10] IPARRAGUIRRE L, MUÑOZ-CULLA M, PRADA-LUENGO I, et al. Circular RNA profiling reveals that circular RNAs from ANXA2 can be used as new biomarkers for multiple sclerosis[J]. Hum Mol Genet, 2017, 26(18): 3564-3572. doi: 10.1093/hmg/ddx243
[11] RAVNIK-GLAVA M, GLAVA D. Circulating RNAs as potential biomarkers in amyotrophic lateral sclerosis[J]. International Journal of Molecular Sciences, 2020, 21(5): 1714. doi: 10.3390/ijms21051714
[12] 北京医学会医学遗传学分会, 北京罕见病诊疗与保障学会. 脊髓性肌萎缩症遗传学诊断专家共识[J]. 中华医学杂志, 2020, 100(40): 3130-3140. doi: 10.3760/cma.j.cn112137-20200803-02267 [13] 杨东铃, 阮毅燕. 脊髓性肌萎缩症治疗研究进展[J]. 中国当代儿科杂志, 2022, 24(2): 204-209. https://www.cnki.com.cn/Article/CJFDTOTAL-DDKZ202202014.htm [14] SINGH R N, SEO J, SINGH N N. RNA in spinal muscular atrophy: therapeutic implications of targeting[J]. Expert Opin Ther Targets, 2020, 24(8): 731-743. doi: 10.1080/14728222.2020.1783241
[15] LI Y, ZHENG Q, BAO C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis[J]. Cell Res, 2015, 25(8): 981-984. doi: 10.1038/cr.2015.82
[16] OJHA R, NANDANI R, CHATTERJEE N, et al. Emerging Role of Circular RNAs as potential biomarkers for the diagnosis of human diseases[J]. Adv Exp Med Biol, 2018, 1087: 141-157.
[17] VEA A, LLORENTE-CORTES V, DE GONZALO-CALVO D, et al. Circular RNAs in blood[J]. Adv Exp Med Biol, 2018, 1087: 119-130.
[18] JIANG M, LASH G E, ZHAO X, et al. CircRNA-0004904, CircRNA-0001855, and PAPP-A: potential novel biomarkers for the prediction of preeclampsia[J]. Cell Physiol Biochem, 2018, 46(6): 2576-2586. doi: 10.1159/000489685
[19] 盛磊, 林慧, 仇妮, 等. 翠云草总黄酮经环状RNA circ_0006528通路抑制结直肠癌恶性生物学行为研究[J]. 实用临床医药杂志, 2022, 26(4): 106-113. doi: 10.7619/jcmp.20213680 [20] 刘德慧, 严玉兰. 环状RNA在肺癌诊断及预后中的研究进展[J]. 实用临床医药杂志, 2021, 25(13): 124-128. doi: 10.7619/jcmp.20211851 [21] CHEN T H. Circulating microRNAs as potential biomarkers and therapeutic targets in spinal muscular atrophy[J]. Ther Adv Neurol Disord, 2020, 13: 1756286420979954.
[22] ABIUSI E, INFANTE P, CAGNOLI C, et al. SMA-miRs (miR-181a-5p, -324-5p, and -451a) are overexpressed in spinal muscular atrophy skeletal muscle and serum samples[J]. Elife, 2021, 10: e68054. doi: 10.7554/eLife.68054
[23] KYE M J, GONÇALVES IDO C. The role of miRNA in motor neuron disease[J]. Front Cell Neurosci, 2014, 8: 15.
[24] BONANNO S, MARCUZZO S, MALACARNE C, et al. Circulating MyomiRs as potential biomarkers to monitor response to nusinersen in pediatric SMA patients[J]. Biomedicines, 2020, 8(2): 21. doi: 10.3390/biomedicines8020021
[25] CATAPANO F, ZAHARIEVA I, SCOTO M, et al. Altered levels of microRNA-9, -206, and -132 in spinal muscular atrophy and their response to antisense oligonucleotide therapy[J]. Mol Ther Nucleic Acids, 2016, 5(7): e331.
[26] SISON S L, PATITUCCI T N, SEMINARY E R, et al. Astrocyte-produced miR-146a as a mediator of motor neuron loss in spinal muscular atrophy[J]. Hum Mol Genet, 2017, 26(17): 3409-3420. doi: 10.1093/hmg/ddx230
[27] 张梦雅. 基于RNA测序的SMN1基因缺失型脊髓性肌肉萎缩症的可变剪接差异性分析[J]. 国际检验医学杂志, 2021, 42(23): 5-5. https://www.cnki.com.cn/Article/CJFDTOTAL-GWSQ202123009.htm [28] GODENA V K, NING K. Phosphatase and tensin homologue: a therapeutic target for SMA[J]. Signal Transduct Target Ther, 2017, 2: 17038. doi: 10.1038/sigtrans.2017.38
[29] KANNAN A, BHATIA K, BRANZEI D, et al. Combined deficiency of senataxin and DNA-PKcs causes DNA damage accumulation and neurodegeneration in spinal muscular atrophy[J]. Nucleic Acids Res, 2018, 46(16): 8326-8346. doi: 10.1093/nar/gky641
[30] TAKAKU M, TSUJITA T, HORIKOSHI N, et al. Purification of the human SMN-GEMIN2 complex and assessment of its stimulation of RAD51-mediated DNA recombination reactions[J]. Biochemistry, 2011, 50(32): 6797-805. doi: 10.1021/bi200828g
[31] TAY S H, ELLIEYANA E N, LE Y, et al. A novel zebrafish model for intermediate type spinal muscular atrophy demonstrates importance of Smn for maintenance of mature motor neurons[J]. Hum Mol Genet, 2021, 30(24): 2488-2502. doi: 10.1093/hmg/ddab212
-
期刊类型引用(2)
1. 李颖颖,张毅. 宫颈环形电切术与冷刀锥切术对高级别宫颈上皮内瘤变患者妊娠结局的影响. 中国临床研究. 2021(05): 624-626 .  百度学术
百度学术
2. 杨春红. 宫颈上皮内瘤变的诊治进展研究. 系统医学. 2019(12): 195-198 .  百度学术
百度学术
其他类型引用(1)




 下载:
下载:


 苏公网安备 32100302010246号
苏公网安备 32100302010246号

