Screening of genetic markers for diagnosis of nasopharyngeal carcinoma based on machine learning algorithm
-
摘要:目的
基于最小绝对收缩和选择算子(LASSO)算法与支持向量机递归特征消除(SVM-RFE)算法筛选用于鼻咽癌(NPC)诊断的特征基因标志物。
方法从GEO数据库下载基因表达微阵列数据集GSE53819、GSE13597作为训练集,从GTEx数据库、ICGC数据库分别下载转录组测序数据集GTEx-NPC、ICGC-NPC作为训练集、验证集。通过基因表达差异分析筛选NPC相关差异表达基因(DEGs),再通过LASSO算法和SVM-RFE算法分别筛选3个训练集中的NPC诊断特征基因。结合外部验证集,通过受试者工作特征(ROC)曲线的曲线下面积(AUC)评估特征基因对NPC的诊断效能。
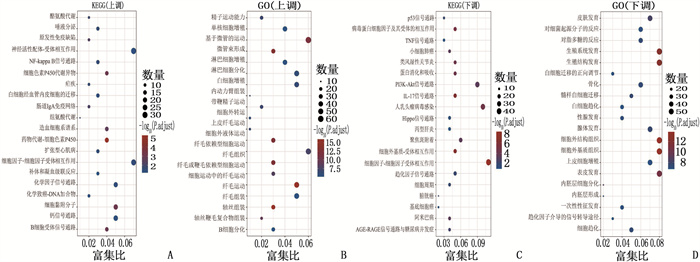
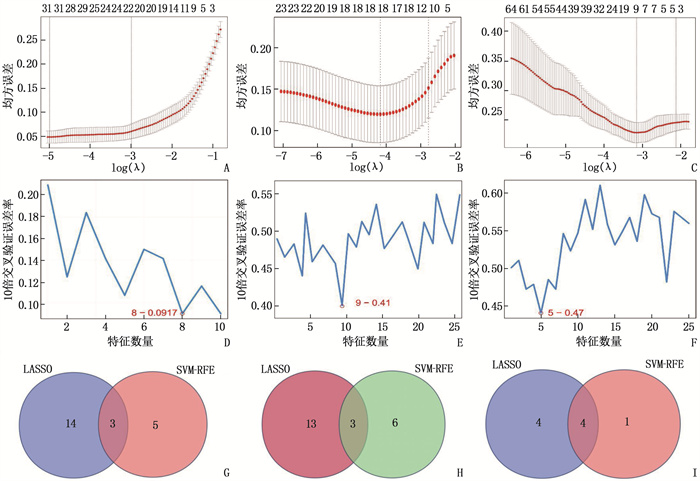
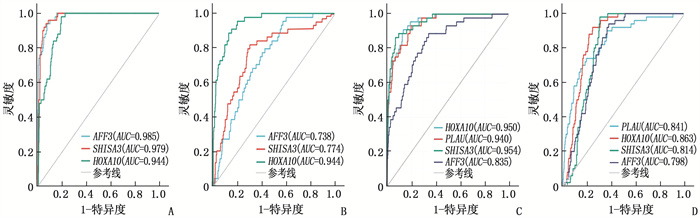
结果本研究共筛选出582个NPC相关DEGs,包括156个高表达DEGs和426个低表达DEGs;基于LASSO算法与SVM-RFE算法,GSE53819、GSE13597、GTEx-NPC数据集均筛选出3个关键诊断特征基因HOXA10、AFF3、SHISA3,且GTEx-NPC数据集另有1个特征基因PLAU;ROC曲线分析结果显示,特征基因HOXA10、AFF3、SHISA3、PLAU在各数据集中诊断NPC的AUC均大于0.7,具有良好的诊断效能。
结论基于LASSO算法和SVM-RFE算法可筛选出4个潜在的NPC诊断特征基因标志物,且外部验证结果显示这些基因标志物在诊断NPC方面具有良好效能,这为NPC的早期诊断和相关基因的分子机制研究提供了有价值的参考。
Abstract:ObjectiveTo screen genetic markers for diagnosis of nasopharyngeal carcinoma (NPC) by the Least Absolute Shrinkage and Selection Operator (LASSO) and Support Vector Machine Recursive Feature Elimination (SVM-RFE) algorithms.
MethodsMicroarray data sets including GSE53819 and GSE13597 of gene expression were downloaded from the GEO database, and transcriptome sequencing data sets including GTEx database and ICGC-NPC database were downloaded as training set and verification set. Differentially expressed genes (DEGs) related to NPC were identified through gene expression differential analysis. Subsequently, LASSO regression and SVM-RFE were used to screen diagnostic feature genes for NPC in three data sets. Finally, an external validation set was used to evaluate the predictive performance of these diagnostic genes by the area under the curve (AUC) of the receiver operating characteristic (ROC) curve.
ResultsA total of 582 DEGs related to NPC were identified, including 156 high expression DEGs and 426 low expression DEGs. Three diagnostic feature genes including HOXA10, AFF3 and SHISA3 were identified by LASSO regression algorithm and SVM-RFE algorithm in the microarray data set. Besides, there was another characteristic gene namely PLAU in the GTEx-NPC dataset. ROC curve analysis results showed that the AUC values of characteristic genes such as HOXA10, AFF3, SHISA3 and PLAU in the diagnosis of NPC in all data sets were greater than 0.7, showing good diagnostic efficacy.
ConclusionFour potential diagnostic feature gene markers for NPC based on LASSO and SVM-RFE algorithm are identified, and they provide valuable references for the diagnosis of NPC, showing a valuable reference for the early diagnosis of NPC and the study of the molecular mechanism of related genes.
-
-
图 3 LASSO算法和SVM-RFE算法筛选关键特征基因的可视化分析图和特征基因交集韦恩图
A: LASSO算法筛选GSE53819数据集的关键特征基因; B: LASSO算法筛选GSE13597数据集的关键特征基因; C: LASSO算法筛选GTEx-NPC数据集的关键特征基因; D: SVM-RFE算法筛选GSE53819数据集的关键特征基因数量; E: SVM-RFE算法筛选GSE13597数据集的关键特征基因数量; F: SVM-RFE算法筛选GTEx-NPC数据集的关键特征基因数量; G: GSE53819数据集不同算法结果的韦恩图; H: GSE13597数据集不同算法结果的韦恩图; I: GTEx-NPC数据集不同算法结果的韦恩图。
表 1 训练集和验证集中特征基因对NPC的诊断效能
数据集 特征基因 曲线下面积 95%置信区间 GSE53819 HOXA10 0.950 0.924~0.975 SHISA3 0.979 0.965~0.993 AFF3 0.985 0.957~1.000 GSE13597 HOXA10 0.944 0.852~1.000 SHISA3 0.774 0.702~0.847 AFF3 0.738 0.675~0.802 GTEx-NPC HOXA10 0.950 0.912~0.986 SHISA3 0.954 0.928~0.992 AFF3 0.835 0.796~0.861 PLAU 0.940 0.902~0.968 ICGC-NPC HOXA10 0.863 0.822~0.894 SHISA3 0.814 0.785~0.843 AFF3 0.798 0.747~0.835 PLAU 0.841 0.801~0.917 -
[1] 周溢, 杨丽, 张妍欣, 等. 鼻咽癌幸存者经济毒性现状及影响因素分析[J]. 军事护理, 2023(1): 15-18. [2] 薛飞, 张婷, 王锐, 等. 鼻咽癌的临床特征及诊断治疗进展[J]. 医学研究生学报, 2022, 35(11): 1213-1218. https://www.cnki.com.cn/Article/CJFDTOTAL-JLYB202211019.htm [3] 吴师雄, 谢静, 方佳宇, 等. 生物信息学方法筛选鼻咽癌的7个关键基因[J]. 武汉大学学报: 医学版, 2022, 43(2): 257-261. https://www.cnki.com.cn/Article/CJFDTOTAL-HBYK202202016.htm [4] 赵琳, 何章彪, 张欣, 等. 利用生物信息学分析鼻咽癌关键基因和信号通路[J]. 中国老年学杂志, 2021, 41(7): 1486-1490. https://www.cnki.com.cn/Article/CJFDTOTAL-ZLXZ202107046.htm [5] ZHANG H, ZOU X, WU L R, et al. Identification of a 7-microRNA signature in plasma as promising biomarker for nasopharyngeal carcinoma detection[J]. Cancer Med, 2020, 9(3): 1230-1241. doi: 10.1002/cam4.2676
[6] GAO P, LU W H, HU S S, et al. Differentially infiltrated identification of novel diagnostic biomarkers associated with immune infiltration in nasopharyngeal carcinoma[J]. Dis Markers, 2022, 2022: 3934704.
[7] 王静娴, 赵芃, 李业棉, 等. 高维生物医学数据变量筛选方法的模拟研究[J]. 西安交通大学学报: 医学版, 2021, 42(4): 628-632. https://www.cnki.com.cn/Article/CJFDTOTAL-XAYX202104027.htm [8] LIN X H, LI C, ZHANG Y H, et al. Selecting feature subsets based on SVM-RFE and the overlapping ratio with applications in bioinformatics[J]. Molecules, 2017, 23(1): 52. doi: 10.3390/molecules23010052
[9] 李慧, 曹博雅, 任璐彤, 等. 基于网络药理学的治伤风颗粒治疗感冒的作用机制探讨[J]. 实用临床医药杂志, 2021, 25(12): 18-23, 41. doi: 10.7619/jcmp.20211607 [10] 盛福梅, 连旭, 韩崇旭. 甲状腺癌差异表达基因的生物信息学分析[J]. 实用临床医药杂志, 2021, 25(10): 1-5, 10. doi: 10.7619/jcmp.20211192 [11] 欧阳天斌. 鼻咽癌患者调强放射治疗后鼻窦炎的临床特征分析[J]. 中国眼耳鼻喉科杂志, 2023, 23(1): 44-48. https://www.cnki.com.cn/Article/CJFDTOTAL-YRBH202301009.htm [12] 陈海珍, 陈建国, 王高仁, 等. 南通市319例鼻咽癌住院患者临床资料分析[J]. 实用肿瘤学杂志, 2022, 36(5): 411-416. https://www.cnki.com.cn/Article/CJFDTOTAL-SYZL202205004.htm [13] GARCIA-MAGARIÑOS M, ANTONIADIS A, CAO R, et al. Lasso logistic regression, GSoft and the cyclic coordinate descent algorithm: application to gene expression data[J]. Stat Appl Genet Mol Biol, 2010, 9: 76-104.
[14] NAN S, SUN L, CHEN B, et al. Density-dependent quantized least squares support vector machine for large data sets[J]. IEEE Trans Neural Netw Learn Syst, 2017, 28(1): 94-106.
[15] DALIRI M R. Feature selection using binary particle swarm optimization and support vector machines for medical diagnosis[J]. Biomed Tech: Berl, 2012, 57(5): 395-402.
[16] DING X J, YANG F, MA F M. An efficient model selection for linear discriminant function-based recursive feature elimination[J]. J Biomed Inform, 2022, 129: 104070.
[17] GONG D, ZHU H, ZENG L, et al. Overexpression of HOXA10 promotes the growth and metastasis of nasopharyngeal carcinoma[J]. Exp Biol Med: Maywood, 2021, 246(23): 2454-2462.
[18] CHEN Z, GONG Q, LI D, et al. CircKIAA0368 promotes proliferation, migration, and invasion by upregulating HOXA10 in nasopharyngeal carcinoma[J]. Am J Rhinol Allergy, 2022, 36(5): 615-627.
[19] ZHANG J, LI Y Q, GUO R, et al. Hypermethylation of SHISA3 promotes nasopharyngeal carcinoma metastasis by reducing SGSM1 stability[J]. Cancer Res, 2019, 79(4): 747-759.
[20] ZENG Y, ZHANG X, LI F, et al. AFF3 is a novel prognostic biomarker and a potential target for immunotherapy in gastric cancer[J]. J Clin Lab Anal, 2022, 36(6): e24437.
[21] LI Z X, CHEN C H, WANG J C, et al. Overexpressed PLAU and its potential prognostic value in head and neck squamous cell carcinoma[J]. PeerJ, 2021, 9: e10746.
[22] DONG Y L, SUN Y, HUANG Y L, et al. Depletion of MLKL inhibits invasion of radioresistant nasopharyngeal carcinoma cells by suppressing epithelial-mesenchymal transition[J]. Ann Transl Med, 2019, 7(23): 741.
[23] 陈彦竹, 何倩, 马宏志, 等. PI3K-Akt/mTOR/AMPK通路基因突变与鼻咽癌疗效及预后的关系[J]. 中南大学学报: 医学版, 2022, 47(2): 165-173. https://www.cnki.com.cn/Article/CJFDTOTAL-HNYD202202003.htm [24] FAN X Q, XIE X N, YANG M, et al. YBX3 mediates the metastasis of nasopharyngeal carcinoma via PI3K/AKT signaling[J]. Front Oncol, 2021, 11: 617621.
-
期刊类型引用(5)
1. 李晖,唐丽华,王敏,宋红花,宋娜,闫克鹏. 消除合并重排简化管理联合风险评估对机械通气中降低多重耐药菌感染发生率的效果研究. 中国医学装备. 2025(02): 99-103 .  百度学术
百度学术
2. 张倩倩,李璐,韩芸. 张家口市某三甲医院急诊科重症患者医院感染现状及病原菌分布情况. 华南预防医学. 2025(02): 222-225 .  百度学术
百度学术
3. 陈晖,张莉,陆晨,蔡新娣. 脑出血术后多重耐药感染风险的诺模图预测模型的构建与验证. 实用临床医药杂志. 2024(08): 45-49+54 .  本站查看
本站查看
4. 章倩影,施英英. 基于人本思想探讨中医肿瘤专科人文病房的多元化构建措施. 中医药管理杂志. 2024(09): 215-217 .  百度学术
百度学术
5. 王浩,代凯利,周记,周政,郑子祥,张静. 中国ICU成人病人多重耐药菌感染影响因素的Meta分析. 循证护理. 2024(22): 3996-4003 .  百度学术
百度学术
其他类型引用(1)





 下载:
下载:
 苏公网安备 32100302010246号
苏公网安备 32100302010246号

