Construction and validation of prognostic risk model for patients with hepatocellular carcinoma based on bioinformatics analysis
-
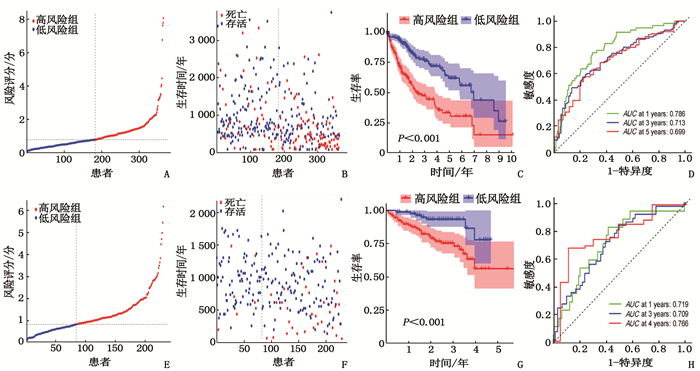
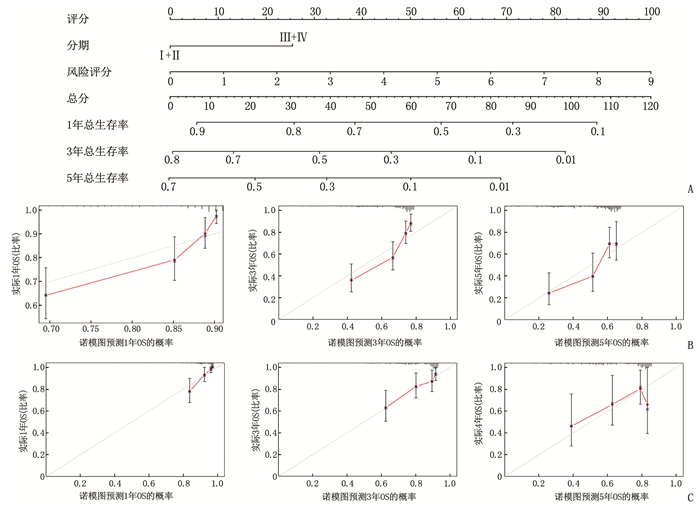
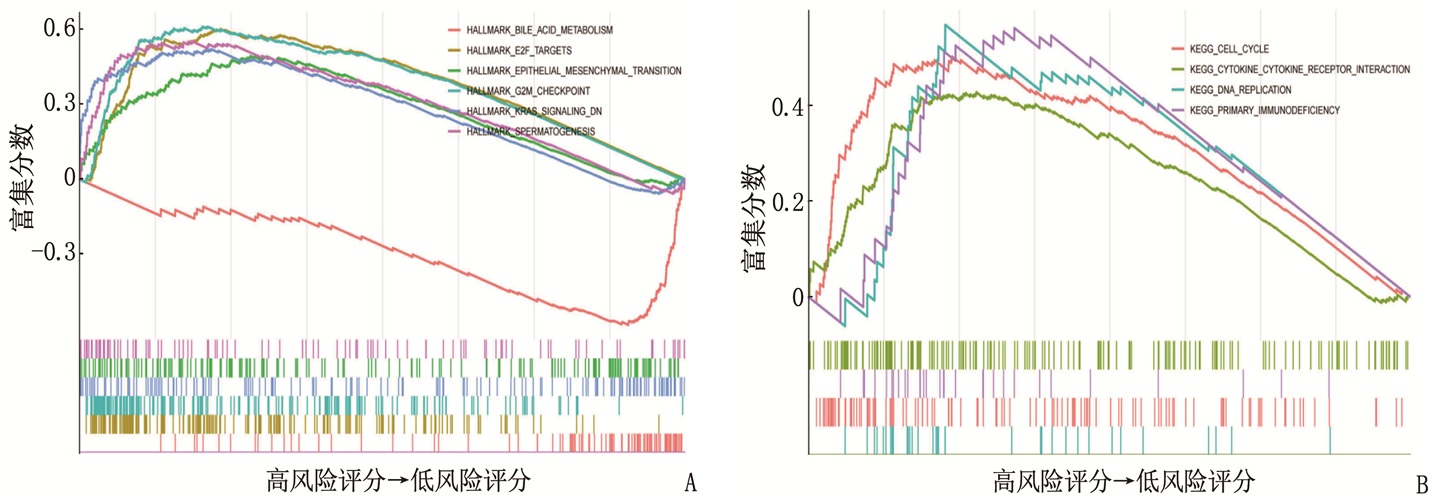
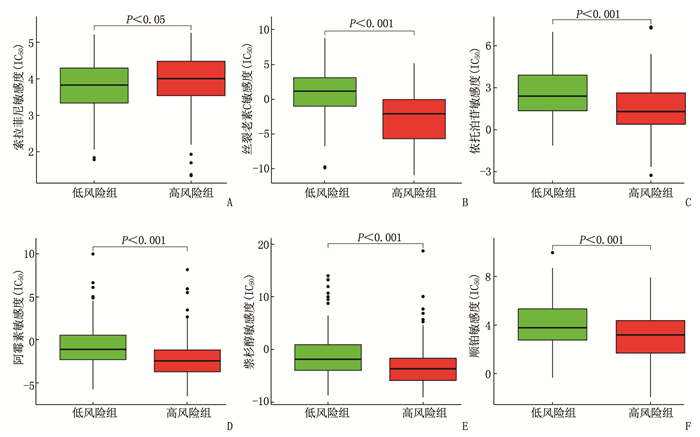
摘要:目的 利用公共数据库构建用于临床治疗肝细胞癌(HCC)的预后风险模型。方法 分别从癌症基因组图谱(TCGA)和国际癌症基因组联盟(ICGC)下载HCC以及癌旁正常组织的mRNA表达数据及临床信息。在TCGA队列中筛选与总生存期(OS)相关的差异表达基因(DEGs), 从中抽取2个或3个mRNAs构成一个组合, 对所有组合进行Cox风险比例回归模型构建。通过受试者工作特征(ROC)曲线的曲线下面积(AUC)确定最优基因组合,并进行基于ICGC队列的外部验证; 以TCGA队列的风险评分中位值将患者分为高风险组与低风险组,进行基因集富集分析(GSEA), 并通过pRRophetic R软件包预测HCC患者使用索拉非尼、丝裂霉素、依托泊苷、阿霉素、紫杉醇和顺铂的相对半抑制浓度(IC50)。结果 该预后风险模型预测TCGA队列的1、3、5年OS的ROC的AUC分别是0.786、0.713、0.699, 预测ICGC队列的1、3、4年OS的ROC的AUC分别为0.719、0.709、0.766。GSEA表明高风险组患者细胞周期相关通路被激活,胆汁酸代谢被抑制。索拉非尼在低风险组的IC50低于高风险组,而细胞周期相关化疗药物在低风险组的IC50高于高风险组,差异均有统计学意义(P < 0.05)。结论 本研究建立并验证了HCC预后风险模型,为HCC患者个体化诊疗方案的制订提供参考依据。Abstract:Objective To construct a prognostic risk model for clinical treatment of hepatocellular carcinoma (HCC) based on public databases.Methods The mRNA expression data and clinical information of HCC and adjacent normal tissues were downloaded from The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC). Differentially expressed genes (DEGs) related to overall survival (OS) were screened in the TCGA cohort, 2 or 3 mRNAs were selected to form a combination, and Cox risk proportional regression model was constructed for all combinations. The optimal gene combination was determined by the area under the curve (AUC) of the receiver operating characteristic (ROC) curve, and external validation based on ICGC cohort was carried out. The patients were divided into high-risk group and low-risk group according to the median risk score of TCGA cohort, gene set enrichment analysis (GSEA) was performed, and the relative half-inhibitory concentrations (IC50) of sorafenib, mitomycin, etoposide, adriamycin, paclitaxel and cisplatin in HCC patients were predicted by pRRophetic R software package.Results For this prognostic risk model, the AUC of the ROC curve for predicting 1-, 3- and 5-year OS in the TCGA cohort were 0.786, 0.713 and 0.699, respectively, and the AUC of the ROC curve for predicting 1-, 3- and 4-year OS in the ICGC cohort were 0.719, 0.709 and 0.766, respectively. GSEA revealed that cell cycle related pathways were activated and bile acid metabolism was inhibited in the high-risk group. The IC50 of sorafenib in the low-risk group was significantly lower than that in the high-risk group, while the IC50 of cell cycle related chemotherapy drugs in the low-risk group was significantly higher than that in the high-risk group (P < 0.05).Conclusion This study establishes and verifies the prognostic risk model for HCC, and provides a reference for the formulation of individualized diagnosis and treatment plan for HCC patients.
-
-
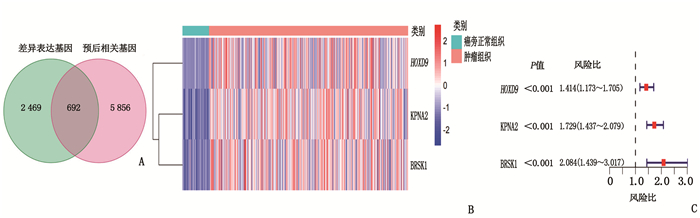
表 1 HCC患者的临床病理资料[n(%)]
临床资料 TCGA队列
(n=365)ICGC队列
(n=231)年龄/岁 61(16, 90) 69(31, 89) 性别 女 119(32.6) 61(26.4) 男 246(67.4) 170(72.6) 肿瘤分级 Ⅰ级 55(15.1) — Ⅱ级 175(47.9) — Ⅲ级 118(32.3) — Ⅳ级 12(3.3) — 未知 5(1.4) — 肿瘤分期 Ⅰ期 170(46.6) 36(15.6) Ⅱ期 84(23.0) 105(45.5) Ⅲ期 83(22.7) 71(30.7) Ⅳ期 4(1.1) 19(8.2) 未知 24(6.6) 0 血管浸润 有 106(29.0) — 无 205(56.2) — 未知 54(14.8) — 甲胎蛋白 ≤200 ng/mL 201(55.1) — > 200 ng/mL 75(20.5) — 未知 89(24.4) — 年龄以中位数(最小值,最大值)表示。 表 2 风险评分与HCC患者临床病理资料的相关性
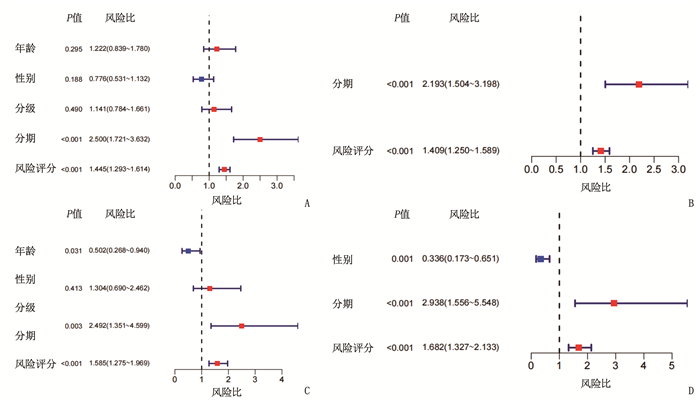
临床资料 TCGA队列 ICGC队列 高风险组 低风险组 P 高风险组 低风险组 P 性别 0.80 1.00 女性 58 61 22 39 男性 125 121 62 108 年龄 0.80 0.42 < 60岁 81 84 29 60 ≥60岁 102 98 55 87 肿瘤分级 < 0.05 - 1、2级 134 96 - - 3、4级 47 83 - - 肿瘤分期 < 0.05 < 0.05 Ⅰ、Ⅱ期 141 113 63 78 Ⅲ、Ⅳ期 30 57 21 69 血管浸润 < 0.05 - 无 124 81 - - 有 44 62 - - 甲胎蛋白 < 0.05 - ≤200 ng/mL 122 79 - - >200 ng/mL 27 48 - - -
[1] VILLANUEVA A. Hepatocellular carcinoma[J]. N Engl J Med, 2019, 380(15): 1450-1462. doi: 10.1056/NEJMra1713263
[2] YANG J D, HAINAUT P, GORES G J, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management[J]. Nat Rev Gastroenterol Hepatol, 2019, 16(10): 589-604. doi: 10.1038/s41575-019-0186-y
[3] TORRE L A, BRAY F, SIEGEL R L, et al. Global cancer statistics, 2012[J]. CA Cancer J Clin, 2015, 65(2): 87-108. doi: 10.3322/caac.21262
[4] BROWN Z J, GRETEN T F, HEINRICH B. Adjuvant treatment of hepatocellular carcinoma: prospect of immunotherapy[J]. Hepatology, 2019, 70(4): 1437-1442. doi: 10.1002/hep.30633
[5] HUANG W T, SKANDERUP A J, LEE C G. Advances in genomic hepatocellular carcinoma research[J]. Gigascience, 2018, 7(11): 1-22. http://www.onacademic.com/detail/journal_1000041617495499_6d02.html
[6] DOMINGUEZ D A, WANG X W. Impact of next-generation sequencing on outcomes in hepatocellular carcinoma: how precise are we really[J]. J Hepatocell Carcinoma, 2020, 7: 33-37. doi: 10.2147/JHC.S217948
[7] CARUSO S, O'BRIEN D R, CLEARY S P, et al. Genetics of hepatocellular carcinoma: approaches to explore molecular diversity[J]. Hepatology, 2021, 73(Suppl 1): 14-26. http://www.researchgate.net/publication/348322949_Genetics_of_Hepatocellular_Carcinoma_Approaches_to_Explore_Molecular_Diversity
[8] GEELEHER P, COX N, HUANG R S. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels[J]. PLoS One, 2014, 9(9): e107468. doi: 10.1371/journal.pone.0107468
[9] PAN Y S, CHEN H R, YU J. Biomarkers in hepatocellular carcinoma: current status and future perspectives[J]. Biomedicines, 2020, 8(12): 576. doi: 10.3390/biomedicines8120576
[10] OURA K, MORISHITA A, MASAKI T. Molecular and functional roles of microRNAs in the progression of hepatocellular carcinoma-A review[J]. Int J Mol Sci, 2020, 21(21): 8362. doi: 10.3390/ijms21218362
[11] KIM E, VIATOUR P. Hepatocellular carcinoma: old friends and new tricks[J]. Exp Mol Med, 2020, 52(12): 1898-1907. doi: 10.1038/s12276-020-00527-1
[12] MUINAO T, DEKA BORUAH H P, PAL M. Multi-biomarker panel signature as the key to diagnosis of ovarian cancer[J]. Heliyon, 2019, 5(12): e02826. doi: 10.1016/j.heliyon.2019.e02826
[13] FOUNTZILAS C, KAKLAMANI V G. Multi-gene panel testing in breast cancer management[J]. Cancer Treat Res, 2018, 173: 121-140.
[14] YANG Z C, ZI Q, XU K, et al. Development of a macrophages-related 4-gene signature and nomogram for the overall survival prediction of hepatocellular carcinoma based on WGCNA and LASSO algorithm[J]. Int Immunopharmacol, 2021, 90: 107238. doi: 10.1016/j.intimp.2020.107238
[15] LIU G M, XIE W X, ZHANG C Y, et al. Identification of a four-gene metabolic signature predicting overall survival for hepatocellular carcinoma[J]. J Cell Physiol, 2020, 235(2): 1624-1636. doi: 10.1002/jcp.29081
[16] LIN P, HE R Q, DANG Y W, et al. An autophagy-related gene expression signature for survival prediction in multiple cohorts of hepatocellular carcinoma patients[J]. Oncotarget, 2018, 9(25): 17368-17395. doi: 10.18632/oncotarget.24089
[17] LI G X, XU W Q, ZHANG L, et al. Development and validation of a CIMP-associated prognostic model for hepatocellular carcinoma[J]. EBioMedicine, 2019, 47: 128-141. doi: 10.1016/j.ebiom.2019.08.064
[18] YU J, WU X L, LV M, et al. A model for predicting prognosis in patients with esophageal squamous cell carcinoma based on joint representation learning[J]. Oncol Lett, 2020, 20(6): 387. http://www.ncbi.nlm.nih.gov/pubmed/33193847
[19] YANG Y J, WANG C Y, WEI N D, et al. Identification of prognostic chromatin-remodeling genes in clear cell renal cell carcinoma[J]. Aging (Albany NY), 2020, 12(24): 25614-25642.
[20] HONG W F, LIANG L, GU Y J, et al. Immune-related lncRNA to construct novel signature and predict the immune landscape of human hepatocellular carcinoma[J]. Mol Ther Nucleic Acids, 2020, 22: 937-947. doi: 10.1016/j.omtn.2020.10.002
[21] 陈懿, 李雪, 林文雅, 等. 自噬基因预测肝癌患者长期生存及通路分析[J]. 医学研究杂志, 2021, 50(1): 137-141. https://www.cnki.com.cn/Article/CJFDTOTAL-YXYZ202101031.htm [22] 段万里, 任伟, 邓骞, 等. 基于TCGA数据库的肾癌自噬相关基因预后模型的建立与应用[J]. 现代泌尿外科杂志, 2020, 25(10): 870-875, 889. doi: 10.3969/j.issn.1009-8291.2020.10.003 [23] TABUSE M, OHTA S, OHASHI Y, et al. Functional analysis of HOXD9 in human gliomas and glioma cancer stem cells[J]. Mol Cancer, 2011, 10: 60. doi: 10.1186/1476-4598-10-60
[24] LONG J Y, ZHANG L, WAN X S, et al. A four-gene-based prognostic model predicts overall survival in patients with hepatocellular carcinoma[J]. J Cell Mol Med, 2018, 22(12): 5928-5938. doi: 10.1111/jcmm.13863
[25] LV X P, LI L L, LV L, et al. HOXD9 promotes epithelial-mesenchymal transition and cancer metastasis by ZEB1 regulation in hepatocellular carcinoma[J]. J Exp Clin Cancer Res, 2015, 34: 133. doi: 10.1186/s13046-015-0245-3
[26] CHRISTIANSEN A, DYRSKJOT L. The functional role of the novel biomarker karyopherin α 2 (KPNA2) in cancer[J]. Cancer Lett, 2013, 331(1): 18-23. doi: 10.1016/j.canlet.2012.12.013
[27] GUO X G, WANG Z H, ZHANG J N, et al. Upregulated KPNA2 promotes hepatocellular carcinoma progression and indicates prognostic significance across human cancer types[J]. Acta Biochim Biophys Sin (Shanghai), 2019, 51(3): 285-292. doi: 10.1093/abbs/gmz003
[28] JIANG P, TANG Y Q, HE L, et al. Aberrant expression of nuclear KPNA2 is correlated with early recurrence and poor prognosis in patients with small hepatocellular carcinoma after hepatectomy[J]. Med Oncol, 2014, 31(8): 1-7.
[29] KONIROVA J, OLTOVA J, CORLETT A, et al. Modulated DISP3/PTCHD2 expression influences neural stem cell fate decisions[J]. Sci Rep, 2017, 7: 41597. doi: 10.1038/srep41597
[30] XIE G X, WANG X N, HUANG F J, et al. Dysregulated hepatic bile acids collaboratively promote liver carcinogenesis[J]. Int J Cancer, 2016, 139(8): 1764-1775. doi: 10.1002/ijc.30219
[31] CHEN W B, OU M L, TANG D E, et al. Identification and validation of immune-related gene prognostic signature for hepatocellular carcinoma[J]. J Immunol Res, 2020, 2020: 5494858. http://qikan.cqvip.com/Qikan/Article/Detail?id=7106408960
[32] STACEY D, KAZLAUSKAS A. Regulation of Ras signaling by the cell cycle[J]. Curr Opin Genet Dev, 2002, 12(1): 44-46. doi: 10.1016/S0959-437X(01)00262-3
[33] MATSUDA Y. Molecular mechanism underlying the functional loss of cyclindependent kinase inhibitors p16 and p27 in hepatocellular carcinoma[J]. World J Gastroenterol, 2008, 14(11): 1734-1740. doi: 10.3748/wjg.14.1734
[34] MATSUDA Y, ICHIDA T. p16 and p27 are functionally correlated during the progress of hepatocarcinogenesis[J]. Med Mol Morphol, 2006, 39(4): 169-175. doi: 10.1007/s00795-006-0339-2
[35] GREENBAUM L E. Cell cycle regulation and hepatocarcinogenesis[J]. Cancer Biol Ther, 2004, 3(12): 1200-1207. doi: 10.4161/cbt.3.12.1392
-
期刊类型引用(1)
1. 吴艳,相开放. 甲状腺乳头状癌组织中MTAP、SETD2表达水平及其与临床病理特征的关系. 黑龙江医药. 2023(03): 704-706 .  百度学术
百度学术
其他类型引用(1)




 下载:
下载:
 苏公网安备 32100302010246号
苏公网安备 32100302010246号
